[ad_1]
Selection of RT-RAA amplification primers and HIV-1 RNA nucleic acid goal, improvement of ERASE detection technique for HIV-1 RNA
In this examine, a fast nucleic acid detection assay for HIV-1 RNA was developed primarily based on the ERASE technique for COVID-19 detection established by our collaborator (Fig. 1A). First, HIV-1 RNA extracted from plasma samples was amplified by the RT-RAA expertise for 30 min, the amplified merchandise had been then added to the CRISPR-Cas13a system. Second, within the CIRSPR/Cas13a system, Cas13a-crRNA advanced acknowledged the goal RNA sequence particularly and activated the cleavage exercise of Cas13a. Third, Cas13a sliced the encircling reporter RNA nonspecifically, the outcomes will be measured by a following fluorescence assay or side-flow strips instantly.
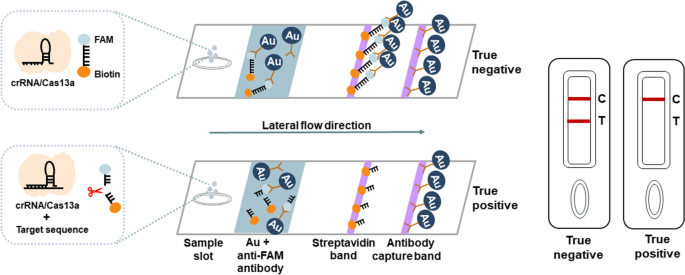
CRISPR-based latera-flow strip for HIV-1 RNA detection. A Schematic of the lateral circulate strip for CRISPR-based detection. CRISPR-Cas13a is mixed with RT-RAA, CRISPR-Cas13a detection is carried out after RT-RAA and the outcomes are learn by fluorescence assay or side-flow strips. B Schematic of RT-RAA primer and crRNA design, RT-RAA upstream primer and downstream primer are mixed in pairs
To develop the RT-RAA-CRISPR-Cas13a assay for detecting HIV-1 RNA, 5086 full genomic sequences of HIV-1 had been downloaded from the HIV community database (http://www.hiv.lanl.gov, launched in 2018) after which in comparison with the genomic sequences in MEGA7.0 and Jalview1.83, 9 pairs of RT-RAA primers and 4 crRNA had been designed (Fig. 1B, Table S1). A extremely conserved sequence (4620-4922 bp) had been screened within the HIV pol area (Table S2) and the plasmid of a HIV-1 customary pressure Psmart-LC (KanR) (GenBank ID: k03455.1) was constructed.
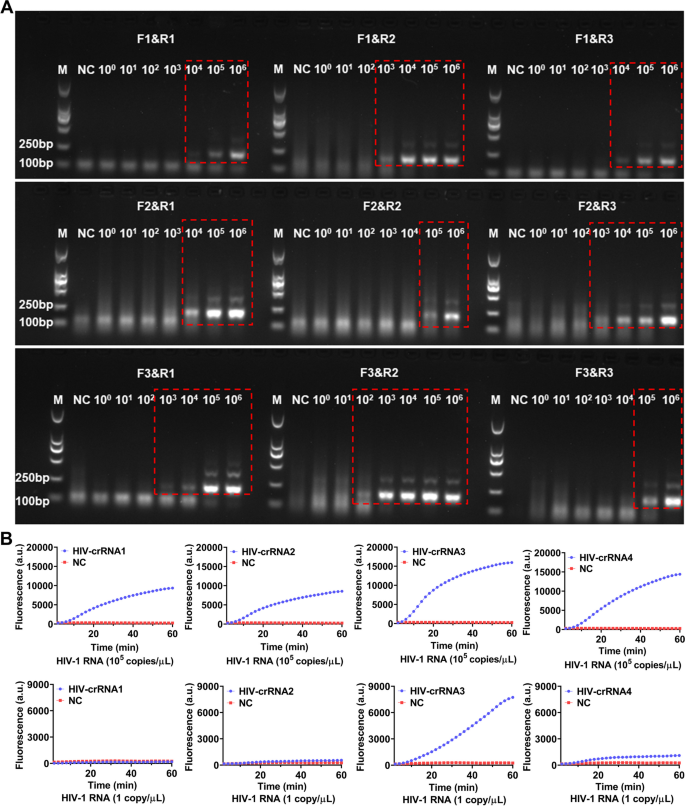
For screening the RT-RAA primers, a serial of diluted HIV-1 RNA templates (1 × 106–1 × 100 copies/μL) had been used for RT-RAA amplification, the amplified merchandise had been then analyzed by electrophoresis. The outcomes confirmed that each one the 9 pairs of RT-RAA primers had been capable of amplify HIV-1 RNA by RT-RAA with the RNA template focus at 1 × 106 copies/μL, solely F3&R2 primers amplified a weaker HIV-1 RNA band (179 bp) with the RNA template focus was 1 × 102 copies/μL, the opposite 8 pairs of RT-RAA primers didn’t amplify any goal RNA by RT-RAA (Fig. 2A). Therefore, F3&R2 primer had been chosen as one of the best RT-RAA primers for this examine. Next, HIV-1 RNA templates with focus of 105 copies/μL and 1 copy/μL had been used for RT-RAA amplification respectively and the amplified merchandise had been then detected by CRISPR-based fluorescence assay for screening crRNA.
Screening of RT-RAA primers and crRNA. A The HIV-1 RNA template was diluted in gradient and amplified individually utilizing 9 pairs of RT-RAA primers. The amplification merchandise are proven within the crimson dashed bins (The uncropped result’s proven in Supplementary Figure S1). M refers to Marker, NC refers to ddH2O. B HIV-1 RNA templates (focus of 1 × 105 copies/μL and 1 × 101 copies/μL) are used for RT-RAA amplification and detected by CRISPR-based fluorescence assay. M refers to Marker, NC refers to unfavorable management
The outcomes confirmed that when the template RNA focus was 105 copies/μL, after 1 h, the typical fluoresence depth of the 4 crRNA reactions, which had been 103.00 ± 80.42 (a.u.), 8296.00 ± 131.10 (a.u.), 15701.00 ± 478.90 (a.u.) and 14145.00 ± 356.80 (a.u.) respectively had been vital increased than the unfavorable management 259.33 (a.u.) (Fig. 2B). When the template RNA focus was 1 copy/μL, after 1 h, the typical fluoresence depth of the 4 crRNA reactions had been 64.00 ± 5.90 (a.u.), 306.30 ± 7.81 (a.u.), 7496.00 ± 37.68 (a.u.) and 801.30 ± 33.52 (a.u.), whereas the unfavorable management was 246 ± 7.93 (a.u.) (Fig. 2B). Therefore, the crRNA-3 which had the best fluorescence depth was chosen because the nucleic acid detection goal for subsequent experiments.
Sensitivity of the CRISPR-based lateral-flow strip assay for detecting HIV-1 RNA
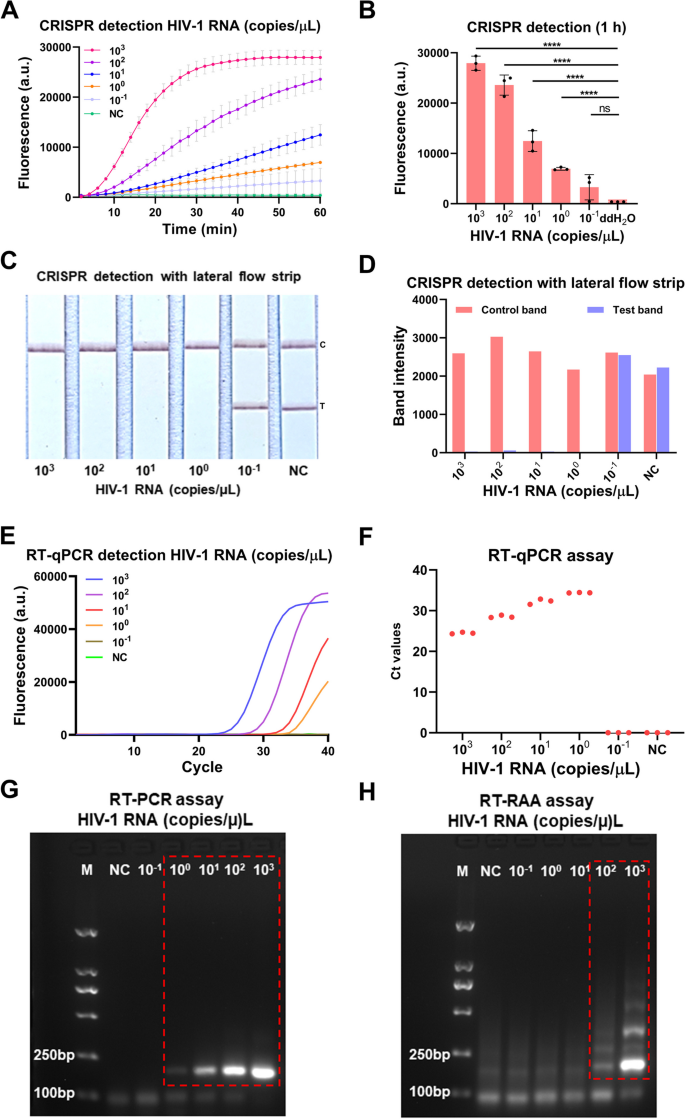
To consider the sensitivity of the our developed CRISPR-based lateral-flow strip assay for detecting HIV-1 RNA (Fig. 3), a serially diluted HIV-1 RNA templates (1 × 103–1 × 10–1 copies/μL) had been measured by CRISPR-based fluorescence assay, CRISPR-based lateral-flow strip assay, RT-qPCR, RT-PCR and RT-RAA amplification. The outcomes confirmed that the sensitivity of the CRISPR-based fluorescence assay reached 1 copy/μL in 10 min (Supplementary Figure S2) and the restrict of detection (LOD) was 1 × 10–1 copies/μL at 1 h (detected twice in 3 impartial repeated trials) (Fig. 4A and B). CRISPR-based lateral-flow strip assay outcomes indicated that when the template RNA focus was 1 copy/μL, solely the C line will be captured however the T line pale away in 2 min, this was acknowledged as a optimistic consequence. When the template RNA focus was 1 × 10–1 copies/μL, each C line and T line will be visualized, this was acknowledged as a unfavorable consequence. So, the sensitivity of CRISPR-based lateral-flow strip assay for detecting HIV-1 RNA was 1 copy/μL (Fig. 4C and D). Similar outcomes had been discovered utilizing RT-qPCR and RT-PCR, each of them had been capable of steadily detect the template RNA at focus of 1 copy/μL (Fig. 4E-G). The LOD of the RT-RAA technique was 1 × 102copies /μL for HIV-1 RNA detection (Fig. 4H).
ERASE lateral circulate strip for CRISPR-based detection. Schematic of the ERASE lateral circulate strip for CRISPR-based detection. Gold-labeled FAM-biotinylated reporter molecules circulate to the check seize band, and redundant gold nanoparticles circulate to the management seize band. Upon recognition of the goal RNA, the crRNA/Cas13a advanced cleaves to the reporter molecule, permitting passage by the check band
Evaluation of the CRISPR-based latera-flow strip for HIV-1 RNA detection. A-B CRISPR-based fluorescence assay for detecting HIV-1 RNA focus at 1 h. C-D CRISPR-based latera-flow strip assay for HIV-1 RNA detection and the band intensities. E–F Distribution of Ct values of RT-qPCR technique and amplification curve of HIV-1 RNA at completely different concentrations. G Agarose gel electrophoresis of HIV-1 RNA with completely different concentrations after RT-PCR (The uncropped result’s proven in Supplementary Figure S3). H Agarose gel electrophoresis of HIV-1 RNA with completely different concentrations after RT-RAA (The uncropped result’s proven in Supplementary Figure S3). NC: Negative management, “****” means P < 0.0001, “***”, “**” means P < 0.005, and “ns” means no vital distinction. All experiments had been carried out in triplicate for knowledge evaluation
The outcomes of evaluating the sensitivity of CRISPR-based lateral-flow strip and CRISPR-based fluorescence assay had been proven in Fig. 4A-D. The fluorescence depth of the HIV-1 RNA with focus of 1 × 103, 1 × 102, 1 × 101, 1 × 100 and 1 × 10–1 copies/μL all elevated because the response time lapsed, after 40 min, the typical fluorescence depth of all of the HIV-1 RNA reactions with completely different focus talked about above, which had been 26921 ± 827.3 (a.u.), 17107 ± 1111 (a.u.), 7031 ± 744.1 (a.u.), 4115 ± 250.0 (a.u.) and 1780 ± 913.3 (a.u.), had been vital increased than the unfavorable management which was 450.00 ± 1.00 (a.u.), no apparent fluorescence indicators had been detected within the group with the goal RNA focus of 1 × 10–1 copies/μL and the unfavorable management group. It indicated that the CRISPR-based lateral-flow strip assay and CRISPR-based fluorescence assay shared the identical sensitivity when detecting HIV-1 RNA.
The serially diluted HIV-1 RNA templates (1 × 103–1 × 10–1 copies/μL) had been used to judge the sensitivity of RT-qPCR, RT-PCR and RT-RAA amplification for detecting HIV RNA (Fig. 4E-H), the sensitivity of the RT-qPCR was 1 × 100 copies/μL and the LOD of the RT-RAA amplification assay was 1 × 102 copies/μL. The outcomes confirmed that the sensitivity of CRISPR-based lateral-flow strip assay was in keeping with the RT-PCR and RT-qPCR, however increased than the RT-RAA amplification assay.
The specificity analysis of CRISPR-based lateral-flow strip assay for detecting HIV-1 RNA
In order to detect the foremost HIV-1 strains worldwide by the CRISPR-Cas13a system, degenerate primers of RT-RAA and crRNA was launched into the HIV-1 pol area (Fig. 5A) to enhance the specificity of the tactic. Then 4 pathogens together with Coxiella burnetiid (Cb), Hepatitis B virus (HBV), Ebola virus (EBOV) and Tick-borne encephalitis virus (TBEV) had been in contrast with HIV-1 by CRISPR-based fluorescence assay and CRISPR-based lateral-flow strip assay on this examine, the outcomes of CRISPR-based fluorescence assay confirmed that the fluorescence depth of HIV-1 reached 3773.00 ± 62.02 (a.u.) after 10 min, it was considerably increased than the fluorescence depth of the nucleic acids of different 4 pathogens (P˂0.05) (Fig. 5B). The fluorescence depth of HIV-1 detection was enhanced quantitatively and stabilized after 1 h, and no vital cross-reaction with different pathogens was noticed (Fig. 5C). The CRISPR-based lateral-flow strip assay confirmed that solely the HIV-1 RNA detection was optimistic (T line pale away and C line appeared), the opposite 4 pathogens had been all unfavorable (Both C line and T line will be noticed) (Fig. 5D and E).
Evaluation of the specificity of CRISPR-Cas13a for HIV-1 detection. A The genome map reveals the location of the goal crRNA and primers on the HIV-1 pol gene. RT-RAA primers have degenerate bases R (A/G) and Y (C/T), that are marked in crimson; the crRNA sequence is marked in yellow. B Fluorescence technique CRISPR-Cas13a detects 5 sorts of pathogen nucleic acids. C Fluorescence worth of various pathogens when CRISPR-Cas13a is detected for 10 min by fluorescence technique. D Test strip technique CRISPR-Cas13a to detect nucleic acids of various pathogens. E Band intensities of every check strip had been quantified utilizing ImageJ. C: Control line, T: Test line, NC: Negative management, “****” means P < 0.0001, the above experiments had been carried out 3 impartial repeat experiments
Evaluation of the CRISPR-based lateral-flow strip assay by scientific samples
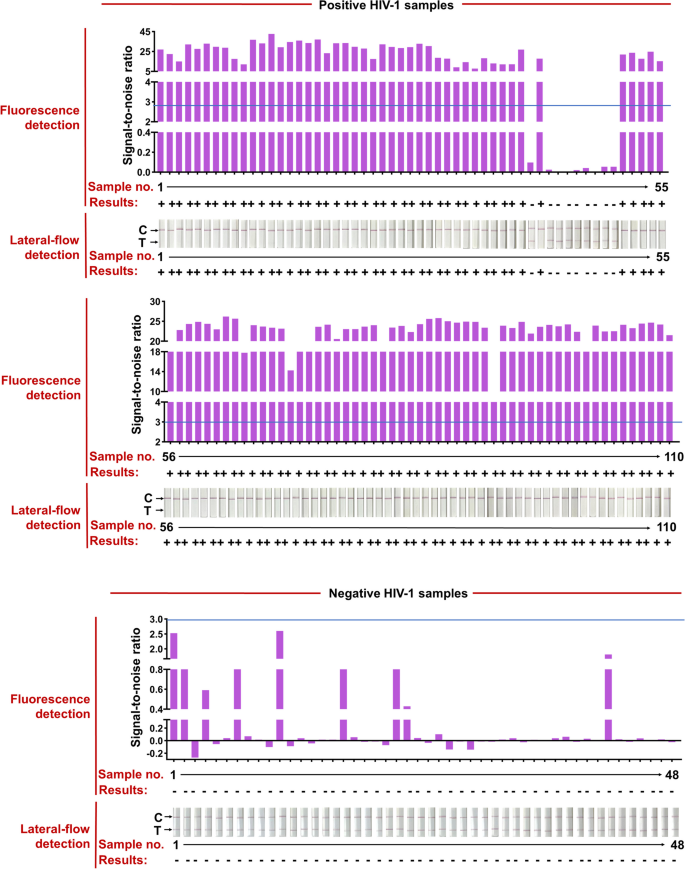
The HIV check kits, RT-qPCR HIV-1 response equipment, used for scientific samples had been bought from the DaAn Gene Co., Ltd. of Sun Yat-sen University. 158 scientific samples from 110 HIV-1-positive sufferers (Ct values assorted from 20 to 40) and 48 HIV-1-negative people had been used to judge the effectiveness of the CRISPR-based lateral-flow strip assay developed on this examine (Fig. 6 and Table 1). All the scientific samples had been detected by the CRISPR-based fluorescence assay and lateral-flow strip CRISPR assay respectively (Supplementary Figure S4).
RT-RAA-CRISPR-Cas13a detected HIV-1 in 158 plasma scientific samples. Positive HIV-1 scientific samples had been categorized in line with their Ct values from RT-qPCR and the entire variety of scientific samples in every Ct classification is proven. Representative lateral-flow strip and fluorescence detection of every Ct bin in addition to HIV-1 unfavorable samples are proven. The full dataset is proven in Supplementary Fig. 1. For lateral-flow strip readings, the disappearance of the coloured check strip, typically mixed with the looks of the coloured management strip, signifies a optimistic HIV-1 consequence. For fluorescence readings, we set a threshold for the signal-to-noise (S/N) ratio of fluorescence depth (blue line) (noise is the fluorescence depth from unfavorable samples carried out in parallel with water as enter), and optimistic outcomes are 3. Bottom row reveals the variety of false unfavorable samples; false unfavorable consequence charges are proven for every Ct bin and for the 2 RT-RAA-CRISPR-Cas13a readout modes
Compared with RT-qPCR, the optimistic fee of CRISPR-based lateral-flow strip assay for detecting HIV-1 RNA was 91.81% (101/110) and the unfavorable fee was 100% (48/48), the general scientific compliance fee was 94.3% with the Kappa worth of 0.872 (P < 0.001), in order the detection outcomes of the CRISPR-based fluorescence assay (Fig. 6, Supplementary Figure S4 and Table 2). The Ct worth of HIV-1 RNA from scientific samples reached 37.32 (112 copies/mL) detected by the CRISPR-based lateral-flow strip assay (Fig. 6, Supplementary S5). For samples with Ct worth much less the 37.32 (viral load extra the 112 copies/mL), the optimistic fee was 100% (100/100) and unfavorable fee was 100% (100/100), and for samples with Ct worth increased than 37.32 (viral load extra the 112 copies/mL), the optimistic fee was 10% (1/10) and unfavorable fee was 100% (48/48) (Fig. 6 and Supplementary S5). Briefly, the outcomes confirmed that the LOD for HIV-1 RNA by the CRISPR-based lateral-flow strip assay was 112 copies/mL, which was extremely in keeping with the scientific analysis.
In order to judge the impact of the CRISPR-based lateral-flow strip assay for detecting completely different HIV-1 subtypes, 60 scientific samples masking three main HIV-1 subtypes in China: subtype 01_AE, 07_BC and subtype B had been collected (20 samples of every subtype) and particular crRNAs had been designed for every subtype (Supplementary Table S3). The HIV-1 RNA was detected by CRISPR-based lateral-flow strip assay and in contrast with RT-qPCR and CRISPR-based fluorescence assay, the outcomes confirmed all of the 60 HIV-1 optimistic samples had been examined optimistic by CRISPR-based lateral-flow strip assay, in addition to by the CRISPR-based fluorescence assay and RT-qPCR (Table 3, Supplementary Figure S5).
[adinserter block=”4″]
[ad_2]
Source link