
[ad_1]

iss057e000185 (10/8/2018) – Biomolecule Sequencer for the BEST experiment floating in entrance of Window 7 within the Cupola module. Earth is within the background. The Biomolecule Sequencer seeks to reveal, for the primary time, that DNA sequencing is possible in an orbiting spacecraft. An area-based DNA sequencer may establish microbes, diagnose ailments and perceive crew member well being, and doubtlessly assist detect DNA-based life elsewhere within the photo voltaic system.
NASA
Bacteria could be recognized by their distinctive organic blueprint, contained inside molecules of deoxyribonucleic acid (DNA).
DNA is made up of 4 base molecules that hyperlink collectively to encode directions for cell progress and conduct. Identifying the order of the bases utilizing the method of DNA sequencing clues researchers in to the identification of the organisms and the way they could behave.
The gear required for DNA sequencing has traditionally been costly and time intensive and has required specialised experience to function, limiting its use in house.
Explore how this expertise has developed to the place researchers can now sequence DNA aboard the International Space Station:
February 1953 – Francis Crick, James Watson, and Rosalind Franklin uncover the double helix construction that makes up DNA.
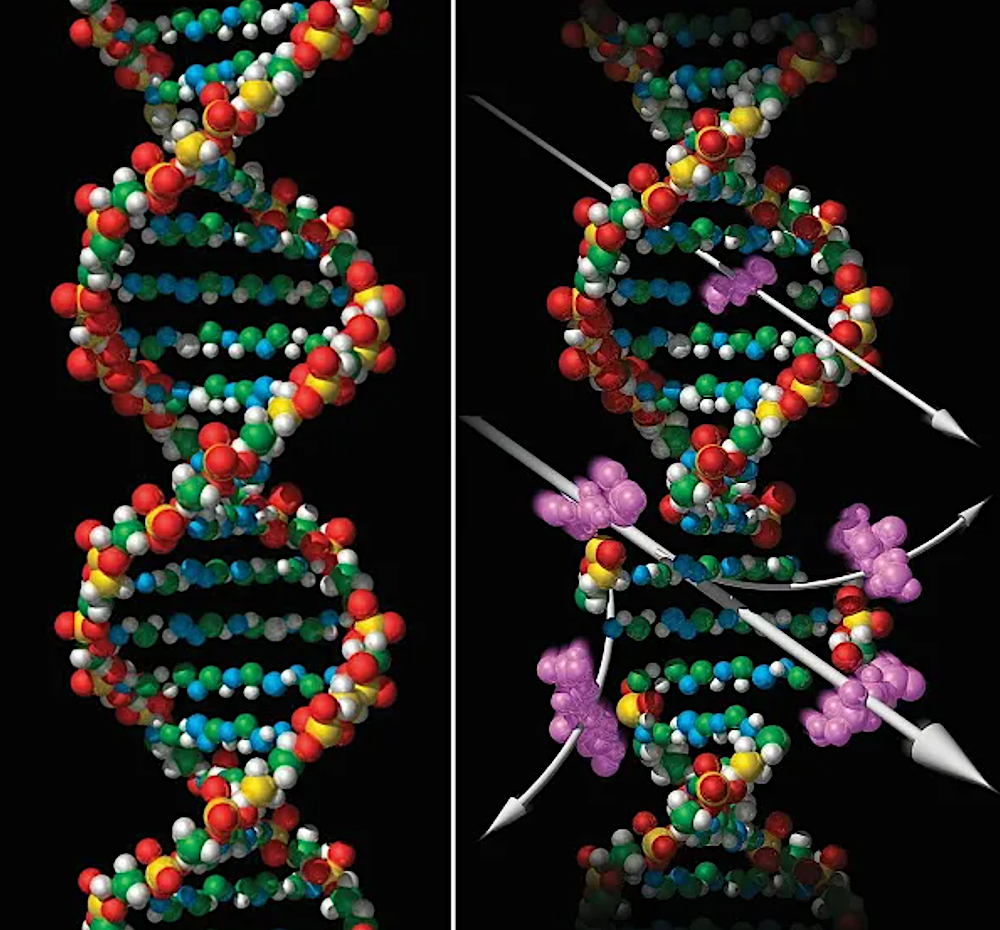
December 1977 – Frederick Sanger develops the primary DNA sequencing methodology to learn the genome of a virus.
July 1995 – First full bacterial genome sequenced (H. influenzae) with shotgun sequencing, which breaks the genome into small fragments which can be sequenced individually utilizing the chain termination methodology, then reassembled.
February 2012 – Oxford Nanopore Technologies debuts the primary nanopore sequencer that makes use of next-generation sequencing (NGS) with the MinION.

April 2016 – As part of the NASA WetLab-2 examine, NASA astronaut Jeff Williams performs the primary RNA isolation in house from E.coli and collects knowledge on the RNA expression ranges within the microbe.
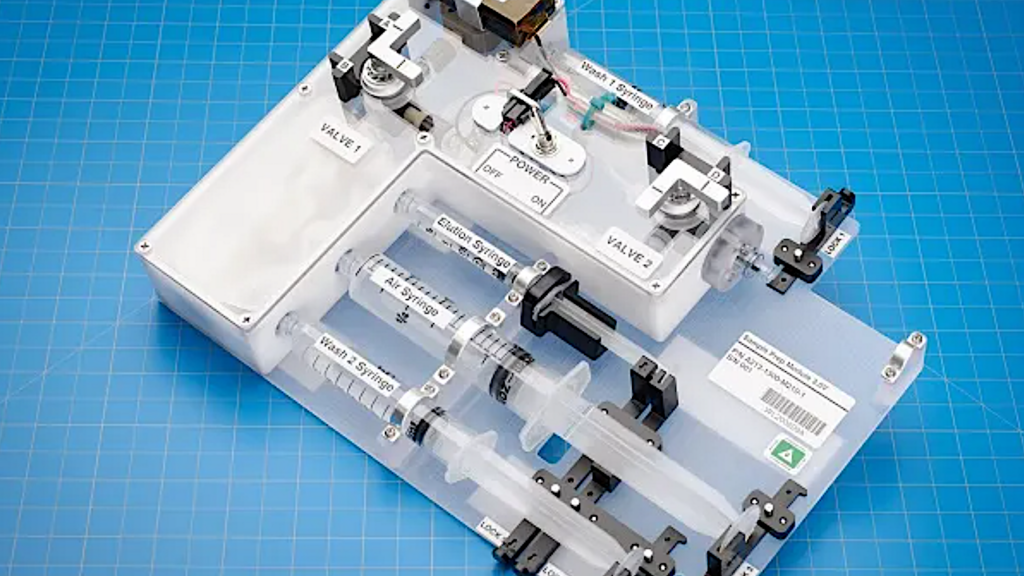
April 2016 – DNA is amplified for the primary time aboard station by ESA (European Space Agency) astronaut Tim Peake utilizing the primary PCR machine despatched to station by firm miniPCR.
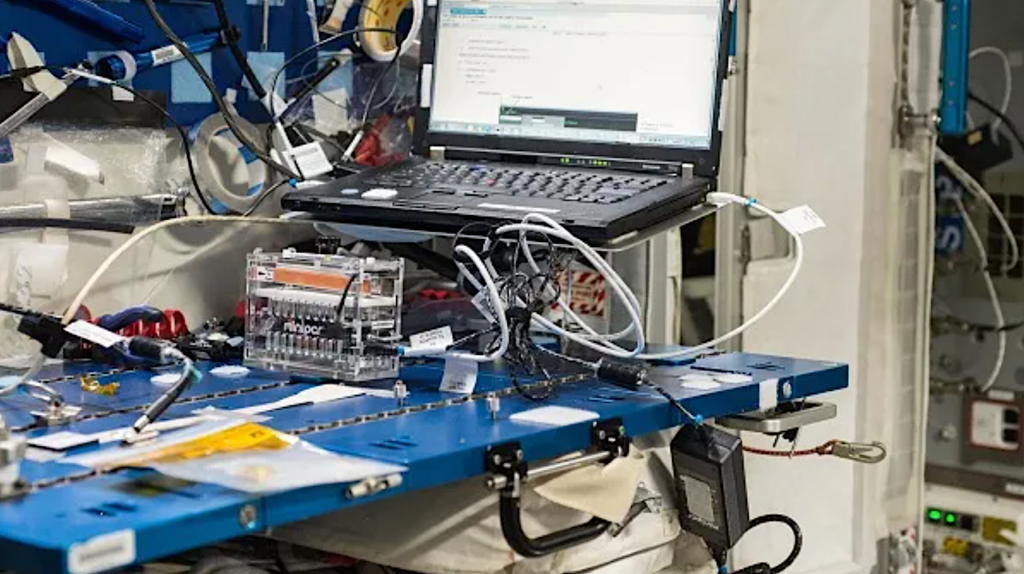
August 2016 – NASA astronaut Kate Rubin sequences DNA in house for the primary time.

August 2017 – NASA astronaut Peggy Whitson combines the miniPCR and MinION, sequencing and figuring out the primary unknown microbe from the station.

View of Genes In Space-3 experiment within the Node 2 module. The Genes in Space-3 experiments reveal methods during which transportable, real-time DNA sequencing can be utilized to assay microbial ecology, diagnose infectious ailments and monitor crew well being aboard the ISS.

August 2018 – NASA astronaut Ricky Arnold demonstrates Biomolecule Extraction and Sequencing Technology (BEST) by utilizing culture-independent strategies to sequence DNA on station for the primary time with a “swab to sequencer” methodology. This course of hurries up the speed of sequencing, now not requiring the time and assets wanted to develop the micro organism previous to evaluation.

May 2019 – NASA astronaut Christina Koch performs the primary CRISPR-Cas9 gene modifying on station, utilizing yeast to imitate the consequences of house radiation on human DNA.

July 31, 2020 – NASA astronaut and Expedition 63 Commander Chris Cassidy works contained in the International Space Station’s Harmony module servicing microbial DNA samples for sequencing and identification.

February 2021 – The crew carried out greater than 800 microbial pattern collections all through station for the 3DMM experiment. Scientists used DNA sequencing and different analyses to assemble the primary complete 3D map of micro organism and bacterial merchandise all through the station.

Jan. 3, 2022 – NASA astronaut and Expedition 66 Flight Engineer Raja Chari sequences DNA from micro organism samples utilizing the BioMole Facility to grasp the microbial atmosphere on the International Space Station.

Sept. 6, 2023 – NASA astronaut and Expedition 69 Flight Engineer Jasmin Moghbeli companies microbe samples for DNA sequencing aboard the International Space Station.

Astrobiology, Genomics, Space Biology,
[adinserter block=”4″]
[ad_2]
Source link