
[ad_1]
Soumya Kannan is a 2021-22 Yang-Tan Center for Molecular Therapeutics Graduate Student Fellow in the lab of MIT Professor Feng Zhang and co-first author with Han Altae-Tran of a study reporting a new class of programmable DNA modifying systems known as OMEGAs. Credit: Caitlin Cunningham
New Programmable Gene Editing Proteins Found Outside of CRISPR Systems
Researchers find RNA-guided enzymes are more diverse and widespread than previously believed.
Within the last decade, scientists have adapted CRISPR systems from microbes into gene editing technology, a precise and programmable system for modifying DNA. Now, scientists at MIT’s McGovern Institute for Brain Research and the Broad Institute of MIT and Harvard have discovered a new class of programmable DNA modifying systems called OMEGAs (Obligate Mobile Element Guided Activity), which may naturally be involved in shuffling small bits of DNA throughout bacterial genomes.
These ancient DNA-cutting enzymes are guided to their targets by small pieces of RNA. While they originated in bacteria, they have now been engineered to work in human cells, suggesting they could be useful in the development of gene editing therapies, particularly as they are small (about 30 percent of the size of Cas9), making them easier to deliver to cells than bulkier enzymes. The discovery, reported on September 9, 2021, in the journal Science, provides evidence that natural RNA-guided enzymes are among the most abundant proteins on Earth, pointing toward a vast new area of biology that is poised to drive the next revolution in genome editing technology.
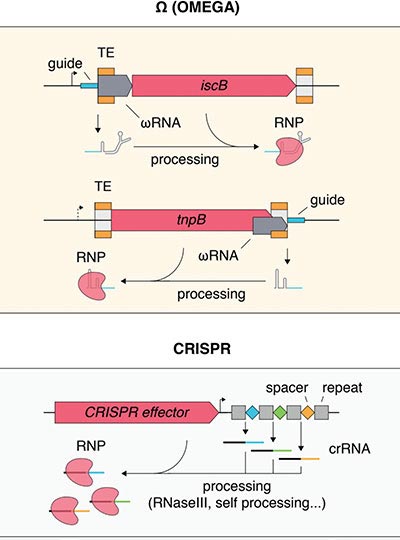
Comparison of Ω (OMEGA) systems with other known RNA-guided systems. In contrast to CRISPR systems, which capture spacer sequences and store them in the locus within the CRISPR array, Ω systems may transpose their loci (or trans-acting loci) into target sequences, converting targets into ωRNA guides. Credit: Courtesy of the researchers
The research was led by McGovern Investigator Feng Zhang, who is the James and Patricia Poitras Professor of Neuroscience at MIT, a Howard Hughes Medical Institute investigator, and a Core Institute Member of the Broad Institute. Zhang’s team has been exploring natural diversity in search of new molecular systems that can be rationally programmed.
“We are super excited about the discovery of these widespread programmable enzymes, which have been hiding under our noses all along,” says Zhang. “These results suggest the tantalizing possibility that there are many more programmable systems that await discovery and development as useful technologies.”
Natural adaptation
Programmable enzymes, particularly those that use an RNA guide, can be rapidly adapted for different uses. For example, CRISPR enzymes naturally use an RNA guide to target viral invaders, but biologists can direct Cas9 to any target by generating their own RNA guide. “It’s so easy to just change a guide sequence and set a new target,” says Soumya Kannan, MIT graduate student in biological engineering and co-first author of the paper. “So one of the broad questions that we’re interested in is trying to see if other natural systems use that same kind of mechanism.”
The first hints that OMEGA proteins might be directed by RNA came from the genes for proteins called IscBs. The IscBs are not involved in CRISPR immunity and were not known to associate with RNA, but they looked like small, DNA-cutting enzymes. The team discovered that each IscB had a small RNA encoded nearby and it directed IscB enzymes to cut specific DNA sequences. They named these RNAs “ωRNAs.”
The team’s experiments showed that two other classes of small proteins known as IsrBs and TnpBs, one of the most abundant genes in bacteria, also use ωRNAs that act as guides to direct the cleavage of DNA.

Zhang lab graduate student Han Altae-Tran is co-author of a recent Science paper on OMEGAS with Soumya Kannan. Credit: Courtesy of the Zhang lab
IscB, IsrB, and TnpB are found in mobile genetic elements called transposons. Han Altae-Tran, MIT graduate student in biological engineering and co-first author on the paper, explains that each time these transposons move, they create a new guide RNA, allowing the enzyme they encode to cut somewhere else.
It’s not clear how bacteria benefit from this genomic shuffling — or whether they do at all. Transposons are often thought of as selfish bits of DNA, concerned only with their own mobility and preservation, Kannan says. But if hosts can “co-opt” these systems and repurpose them, hosts may gain new abilities, as with CRISPR systems that confer adaptive immunity.
IscBs and TnpBs appear to be predecessors of Cas9 and Cas12 CRISPR systems. The team suspects they, along with IsrB, likely gave rise to other RNA-guided enzymes, too — and they are eager to find them. They are curious about the range of functions that might be carried out in nature by RNA-guided enzymes, Kannan says, and suspect evolution likely already took advantage of OMEGA enzymes like IscBs and TnpBs to solve problems that biologists are keen to tackle.
“A lot of the things that we have been thinking about may already exist naturally in some capacity,” says Altae-Tran. “Natural versions of these types of systems might be a good starting point to adapt for that particular task.”
The team is also interested in tracing the evolution of RNA-guided systems further into the past. “Finding all these new systems sheds light on how RNA-guided systems have evolved, but we don’t know where RNA-guided activity itself comes from,” Altae-Tran says. Understanding those origins, he says, could pave the way to developing even more classes of programmable tools.
Reference: “The widespread IS200/IS605 transposon family encodes diverse programmable RNA-guided endonucleases” by Han Altae-Tran, Soumya Kannan, F. Esra Demircioglu, Rachel Oshiro, Suchita P. Nety, Luke J. McKay, Mensur Dlakić, William P. Inskeep, Kira S. Makarova, Rhiannon K. Macrae, Eugene V. Koonin and Feng Zhang, 1 October 2021, Science.
DOI: 10.1126/science.abj6856
This work was made possible with support from the Simons Center for the Social Brain at MIT, the National Institutes of Health and its Intramural Research Program, Howard Hughes Medical Institute, Open Philanthropy, G. Harold and Leila Y. Mathers Charitable Foundation, Edward Mallinckrodt, Jr. Foundation, Poitras Center for Psychiatric Disorders Research at MIT, Hock E. Tan and K. Lisa Yang Center for Autism Research at MIT, Yang-Tan Center for Molecular Therapeutics at MIT, Lisa Yang, Phillips family, R. Metcalfe, and J. and P. Poitras.
[ad_2]
Source link