
[ad_1]
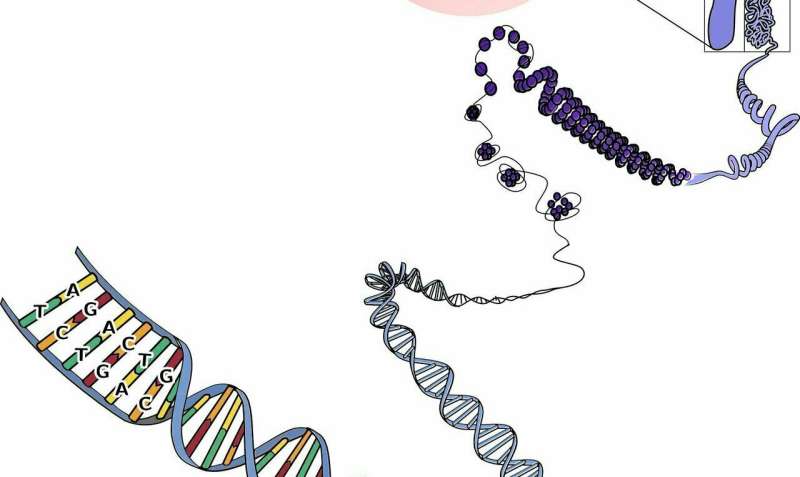
On the journey from gene to protein, a nascent RNA molecule will be reduce and joined, or spliced, in numerous methods earlier than being translated right into a protein. This course of, generally known as different splicing, permits a single gene to encode a number of completely different proteins. Alternative splicing happens in lots of organic processes, like when stem cells mature into tissue-specific cells. In the context of illness, nevertheless, different splicing will be dysregulated. Therefore, it is very important study the transcriptome—that’s, all of the RNA molecules which may stem from genes—to know the foundation explanation for a situation.
However, traditionally it has been tough to “read” RNA molecules of their entirety as a result of they’re normally 1000’s of bases lengthy. Instead, researchers have relied on so-called short-read RNA sequencing, which breaks RNA molecules and sequence them in a lot shorter items—someplace between 200 to 600 bases, relying on the platform and protocol. Computer packages are then used to reconstruct the complete sequences of RNA molecules.
Short-read RNA sequencing may give extremely correct sequencing knowledge, with a low per-base error charge of roughly 0.1% (which means one base is incorrectly decided for each 1,000 bases sequenced). Nevertheless, it’s restricted within the info that it could possibly present because of the quick size of the sequencing reads. In some ways, short-read RNA sequencing is like breaking a big image into many jigsaw items which might be all the identical form and dimension after which making an attempt to piece the image again collectively.
Recently, “long-read” platforms that may sequence RNA molecules over 10,000 bases in size end-to-end have turn into out there. These platforms don’t require RNA molecules to be damaged up earlier than they’re sequenced, however they’ve a a lot greater per-base error charge, sometimes between 5% to twenty%. This well-known limitation has severely hampered the widespread adoption of long-read RNA sequencing. In explicit, the excessive error charge has made it tough to find out the validity of novel, beforehand unknown RNA molecules found in a selected situation or illness.
To circumvent this downside, researchers at Children’s Hospital of Philadelphia (CHOP) have developed a brand new computational device that may extra precisely uncover and quantify RNA molecules from these error-prone long-read RNA sequencing knowledge. The device, known as ESPRESSO (Error Statistics PRomoted Evaluator of Splice Site Options), was reported right now in Science Advances.
“Long-read RNA sequencing is a powerful technology that will allow us to uncover RNA variation in rare genetic diseases and other conditions, like cancer,” stated Yi Xing, Ph.D., director of the Center for Computational and Genomic Medicine at CHOP and senior writer of the research.
“We are probably at an inflection point in how we discover and analyze RNA molecules. The transition from short-read to long-read RNA sequencing represents an exciting technological transformation, and computational tools that reliably interpret long-read RNA sequencing data are urgently needed.”
ESPRESSO can precisely uncover and quantify completely different RNA molecules from the identical gene—generally known as RNA isoforms—utilizing error-prone long-read RNA sequencing knowledge alone. To accomplish that, the computational device compares all lengthy RNA sequencing reads of a given gene to its corresponding genomic DNA, after which makes use of the error patterns of particular person lengthy reads to confidently establish splice junctions—locations the place the nascent RNA molecule has been reduce and joined—in addition to their corresponding full-length RNA isoforms.
By discovering areas of good matches between lengthy RNA sequencing reads and genomic DNA, in addition to borrowing info throughout all lengthy RNA sequencing reads of a gene, the device is ready to establish extremely dependable splice junctions and RNA isoforms, together with people who haven’t been beforehand documented in current databases.
The researchers evaluated the efficiency of ESPRESSO utilizing simulated knowledge and knowledge on actual organic samples. They discovered that ESPRESSO performs higher than a number of presently out there instruments, each by way of discovering RNA isoforms and quantifying them. The researchers additionally generated and analyzed over 1 billion lengthy RNA sequencing reads protecting 30 human tissue varieties and three human cell strains, offering a helpful useful resource for finding out human transcriptome variation on the decision of full-length RNA isoforms.
“ESPRESSO addresses a long-standing problem of long-read RNA sequencing and could usher in new opportunities of discovery,” Dr. Xing stated. “We envision that ESPRESSO will be a useful tool for researchers to explore the RNA repertoire of cells in various biomedical and clinical settings.”
More info:
Yuan Gao et al, ESPRESSO: Robust discovery and quantification of transcript isoforms from error-prone long-read RNA-seq knowledge, Science Advances (2023). DOI: 10.1126/sciadv.abq5072. www.science.org/doi/10.1126/sciadv.abq5072
Provided by
Children’s Hospital of Philadelphia
Citation:
Researchers develop new, extra correct computational device for long-read RNA sequencing (2023, January 20)
retrieved 20 January 2023
from https://phys.org/news/2023-01-accurate-tool-long-read-rna-sequencing.html
This doc is topic to copyright. Apart from any truthful dealing for the aim of personal research or analysis, no
half could also be reproduced with out the written permission. The content material is supplied for info functions solely.
[adinserter block=”4″]
[ad_2]
Source link